Plant-pathogen dynamics
A genomic and transcriptomic co-evolutionary arms race
Fungal disease has devastated plants and animals for millennia, starting from biblical plagues. A pathogen’s success depends on its genetic adaptability, including traits like mutation and horizontal gene transfer: these processes can lead to new effectors, proteins that help pathogens suppress host defences. I first identified fungal effector genes through comparative genomics and subsequently found several up-regulated in infection, alongside resistance proteins in the plant hosts.

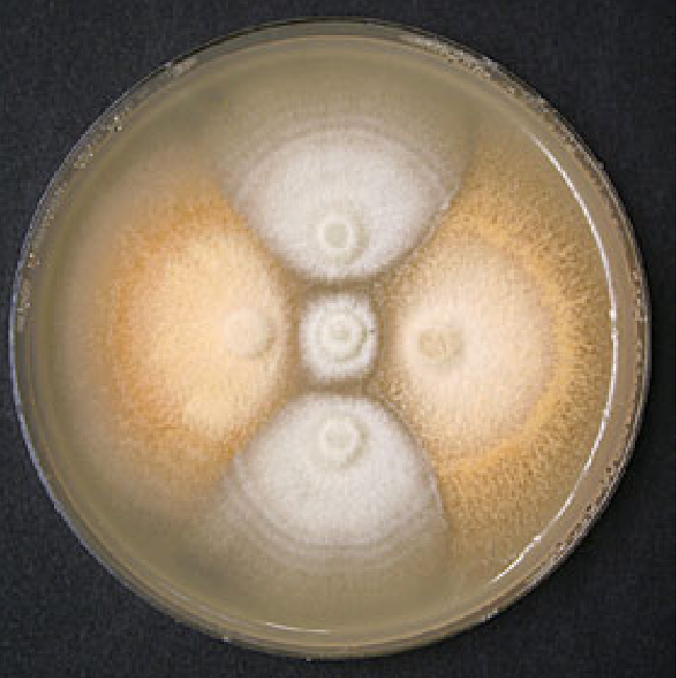
Left: Robusta coffee tree affected by coffee wilt disease in Uganda (Photograph © S. Olal). Right: Strains of Fusarium xylarioides, the coffee wilt disease pathogen, are differentiated by pigmentation on carrot agar: the population infecting robusta coffee are orange; the population infecting arabica coffee are white. The two strains are incompatible, as shown by the absence of perithecia (fruiting bodies) along the contact zones (Photograph © P. Lepoint and H. Maraite).
Representative publications on this topic include:
- Peck, L.D., Llewellyn, T., Bennetot, B., O’Donnell, S., Nowell, R.W., Ryan, M.J., Flood, J., Rodríguez De La Vega, R.C., Ropars, J., Giraud, T., Spanu, P.D. and Barraclough, T.G. (2024) ‘Horizontal transfers between fungal Fusarium species contributed to successive outbreaks of coffee wilt disease’, PLoS Biology 22, 12: e3002480.
- Peck, L.D., Nowell, R.W., Flood, J., Ryan, M.J. and Barraclough, T.G. (2021) ‘Historical genomics reveals the evolutionary mechanisms behind multiple outbreaks of the host-specific coffee wilt pathogen Fusarium xylarioides’, BMC Genomics, 22(1), p. 404.
- Peck, L. (2023) ‘Seventy years of Fusarium wilt-coffee interactions: historical genomics reveals pathogen emergence and divergence’, chapter 4. PhD Thesis. Imperial College London.